Overview¶
In this tutorial, we’ll plot NASA/CNES’s Surface Water and Ocean Topography (SWOT) High Resolution (HR) and Low Resolution (LR) coverage throughout Antarctica, then compare SWOT elevations to NASA’s Ice, Cloud, and Land Elevation Satellite-2 (ICESat-2) over the Bach Ice Shelf (Antarctic Peninsula) to verify that geophysical corrections have been correctly applied.
Visualize SWOT LR and HR coverage over Antarctica¶
Building on the previous SWOT HR data access tutorial where we learned how to find and open SWOT data, we’ll now focus on what’s available for our ICESat-2 comparison. In this step, you’ll create an interactive map of SWOT coverage over Antarctica.
On KML/KMZ Files
The orbit files are hosted in different locations and as different file types; because this tutorial downloads the orbit files directly from the source locations, the HR comes as a KML and LR orbit as a KMZ. A KMZ file is just a zipped version of KML that can typically be read by the same programs designed to read KML files (e.g., Fiona, Google Earth).
Package imports and data paths needed to get started¶
%matplotlib widget
from pathlib import Path
import io, zipfile, tempfile, os
import warnings
import requests
import fiona
import geopandas as gpd
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.colors import to_rgba
from matplotlib.patches import Patch
import cartopy.crs as ccrs
# Quiet noisy driver warnings (optional)
warnings.filterwarnings("ignore", message=".*ERROR parsing kml Style: No id.*", category=RuntimeWarning)# --- Paths ---
# A shared folder on CryoCloud / local machine
DATADIR = Path("/home/jovyan/shared/SWOT/data") # adjust if needed
DATADIR.mkdir(parents=True, exist_ok=True)
# CryoCloud file targets
GL_SHP = DATADIR / "Antarctica_masks" / "scripps_antarctica_polygons_v1.shp"
HR_KML = DATADIR / "hr_Mar2025_below60S.kml" # culled HR KML we’ll create if missing
LR_KMZ = DATADIR / "lr_Sept2015.kmz" # LR orbit
# Source URLs (try HR 2025, fall back to 2024 seasonal if needed)
HR_URLS = [
"https://podaac.jpl.nasa.gov/SWOT-events/swot_science_hr_Mar2025-v10_perPass.kml",
"https://podaac.jpl.nasa.gov/SWOT-events/swot_science_hr_Nov2024-v09-seasonal_perPass.kml",
]
LR_URL = "https://www.aviso.altimetry.fr/fileadmin/documents/missions/Swot/swot_science_orbit_sept2015-v2_10s.kmz"
# Scripps polygons zip (contains multiple shapefile components)
GL_ZIP_URL = "https://doi.pangaea.de/10013/epic.42133.d001"
# Bach Ice Shelf bounding box location in South Polar Stereographic [ESPG: 3031]
bach = [-1920000, 480000, -1740000, 680000]# --- Small helper functions ---
def download_bytes(url: str) -> bytes:
r = requests.get(url, timeout=180)
r.raise_for_status()
return r.content
def ensure_scripps_polygons(gl_shp_path: Path, source_zip_url: str) -> None:
"""Download & extract the Scripps Antarctica polygons if missing."""
if gl_shp_path.exists():
return
gl_shp_path.parent.mkdir(parents=True, exist_ok=True)
print("Downloading Scripps Antarctica polygons…")
content = download_bytes(source_zip_url)
with zipfile.ZipFile(io.BytesIO(content)) as zf:
zf.extractall(gl_shp_path.parent)
print(f"Extracted to: {gl_shp_path.parent}")
def read_all_kml_layers(path: Path, driver="KML") -> gpd.GeoDataFrame:
"""Load all non-empty KML/KMZ layers into a single GeoDataFrame."""
layers = fiona.listlayers(str(path))
frames = []
for lyr in layers:
try:
gdf = gpd.read_file(path, driver=driver, layer=lyr)
if not gdf.empty:
frames.append(gdf)
except Exception as exc:
print(f"Skipped layer '{lyr}': {exc}")
if not frames:
raise RuntimeError(f"No valid layers in {path}")
return gpd.GeoDataFrame(pd.concat(frames, ignore_index=True))
def save_kml(gdf: gpd.GeoDataFrame, out_path: Path) -> None:
"""Save a GeoDataFrame as KML (for future reuse)."""
out_path.parent.mkdir(parents=True, exist_ok=True)
gdf.to_file(out_path, driver="KML")# --- Ensure basemap (Scripps polygons) is available ---
ensure_scripps_polygons(GL_SHP, GL_ZIP_URL)
# Load and prep Scripps polygons
gl_gdf = gpd.read_file(GL_SHP)
# Reproject to EPSG:3031 (Antarctic Polar Stereographic)
gl_gdf_3031 = gl_gdf.to_crs(epsg=3031)
# Split out grounded ice vs ice shelves for nicer styling
name_field = "Id_text" if "Id_text" in gl_gdf_3031.columns else None
if name_field is None:
# Fall back to plotting all polygons uniformly if classification is missing
grounded_3031 = gl_gdf_3031
shelves_3031 = gpd.GeoDataFrame(geometry=[])
else:
grounded_classes = {"Grounded ice or land", "Isolated island", "Ice rise or connected island"}
grounded_3031 = gl_gdf_3031[gl_gdf_3031[name_field].isin(grounded_classes)]
shelves_3031 = gl_gdf_3031[gl_gdf_3031[name_field] == "Ice shelf"]On culling the HR mask
We are able to cull the HR mask to below 60°S because each segment of HR data is discontinuous from other segments even if they are part of the same pass, resulting in the ability to filter for segments using bounding box criteria. Because the LR orbit file is split by pass, each pass contains data below 60°S and cannot easily be culled in the same way. If you run the culling yourself it can take ~23 minutes on CryoCloud.
# --- Ensure SWOT HR coverage (culled to south of 60S) exists in CryoCloud ---
# Note: This process takes 23 min if not already completed
if not HR_KML.exists():
for url in HR_URLS:
try:
# Download HR orbital file into temp
print(f"Trying HR KML: {url}")
with tempfile.TemporaryDirectory() as tdir:
kml_path = Path(tdir) / "hr.kml"
kml_path.write_bytes(download_bytes(url))
hr_all = read_all_kml_layers(kml_path, driver="KML") # with some KMZ/KML may need to use driver="LIBKML"
hr_all = hr_all.assign(miny=hr_all.geometry.bounds.miny)
# Cull below 60 S
hr_south = hr_all[hr_all["miny"] < -60].drop(columns=["miny"])
if "Name" in hr_south.columns:
# Remove nadir coverage to keep only KaRIn's
hr_south = hr_south[~hr_south["Name"].str.contains("Nadir", na=False)]
save_kml(hr_south, HR_KML)
print(f"Saved culled HR KML: {HR_KML}")
break
except Exception as exc:
print(f"Failed HR download/parse from {url}: {exc}")
else:
raise RuntimeError("Could not obtain a usable SWOT HR KML.")
# --- Ensure SWOT LR orbit exists in CryoCloud ---
if not LR_KMZ.exists():
print("Downloading SWOT LR KMZ…")
LR_KMZ.write_bytes(download_bytes(LR_URL))
print(f"Saved: {LR_KMZ}")# --- Load HR/LR coverages and reproject to EPSG:3031 ---
# HR (already culled), stored as KML
hr_gdf = read_all_kml_layers(HR_KML, driver="KML")
hr_3031 = hr_gdf.to_crs(epsg=3031)
# LR (KMZ) — many GDAL/Fiona builds can read KMZ directly with driver="KML"
lr_gdf = read_all_kml_layers(LR_KMZ, driver="KML")
if "Name" in lr_gdf.columns:
lr_gdf = lr_gdf[~lr_gdf["Name"].str.contains("Nadir", na=False)]
lr_3031 = lr_gdf.to_crs(epsg=3031)
len(hr_3031), len(lr_3031)WARNING:fiona._err:CPLE_AppDefined:ERROR parsing kml Style: No id
(194, 584)# --- Plot availability over Antarctica (EPSG:3031) ---
ps71 = ccrs.Stereographic(central_latitude=-90, central_longitude=0, true_scale_latitude=-71)
fig = plt.figure(figsize=(8, 8), dpi=110)
ax = plt.axes(projection=ps71)
# Antarctica basemap: grounded vs shelves
grounded_3031.plot(ax=ax, transform=ps71, color="lightgray", edgecolor="darkgray", linewidth=0.3, zorder=1)
shelves_3031.plot(ax=ax, transform=ps71, color="gainsboro", edgecolor="silver", linewidth=0.25, zorder=2)
# SWOT LR (red), SWOT HR (blue)
# Build colors with different transparency for facecolor and edgecolor
swot_hr_fill = to_rgba("blue", alpha=0.20)
swot_hr_edge = to_rgba("blue", alpha=0.30)
swot_lr_fill = to_rgba("red", alpha=0.06)
swot_lr_edge = to_rgba("red", alpha=0.10)
# Note: Many HR/LR geometries are polygons; we use both facecolor and edgecolor lightly
lr_3031.plot(ax=ax, transform=ps71, color=swot_lr_fill, edgecolor=swot_lr_edge, linewidth=0.2, zorder=3)
hr_3031.plot(ax=ax, transform=ps71, color=swot_hr_fill, edgecolor=swot_hr_edge, linewidth=0.2, zorder=4)
bach_rect = plt.Rectangle((bach[0], bach[1]), bach[2] - bach[0], bach[3] - bach[1],
zorder=5, linewidth=2, edgecolor="white", facecolor="none")
ax.add_patch(bach_rect)
# Crop axes to Antarctica
ax.set_xlim(-3000000, 3000000) # 3000 km2
ax.set_ylim(-3000000, 3000000)
legend_handles = [
Patch(facecolor="gainsboro", edgecolor="silver", label="Ice shelves"),
Patch(facecolor="lightgray", edgecolor="darkgray", label="Grounded ice / land"),
Patch(facecolor=swot_lr_fill, edgecolor=swot_lr_edge, label="SWOT LR"),
Patch(facecolor=swot_hr_fill, edgecolor=swot_hr_edge, label="SWOT HR"),
]
leg = ax.legend(handles=legend_handles, loc="lower left", frameon=True, framealpha=0.9)
for text in leg.get_texts():
text.set_fontsize(10)
ax.set_title("SWOT Availability over Antarctica (N. Hemisphere Summer)", fontsize=14, pad=10)
plt.tight_layout()
plt.show()Compare SWOT and ICESat-2 elevations on Bach Ice Shelf¶
We can see that there is HR coverage over Bach Ice Shelf so we will compare SWOT heights there to ICESat-2.
ICESat-2 is a photon-counting green laser altimeter. We can make this comparison because when measuring snow and ice surface heights, both SWOT (Ka band radar) and ICESat-2 (photon counting green laser) are considered surface sensing instruments. They penetrate to nearly equivalent depths into snow and ice, though Ka band radar likely penetrates millimeters to a few centimeters deeper into dry snow than ICESat-2 does. They are likely more indistinguishable for wet snow. More information about ICEsat-2 data products, mission, and tutorials is available in the ICESat-2 Cookbook.
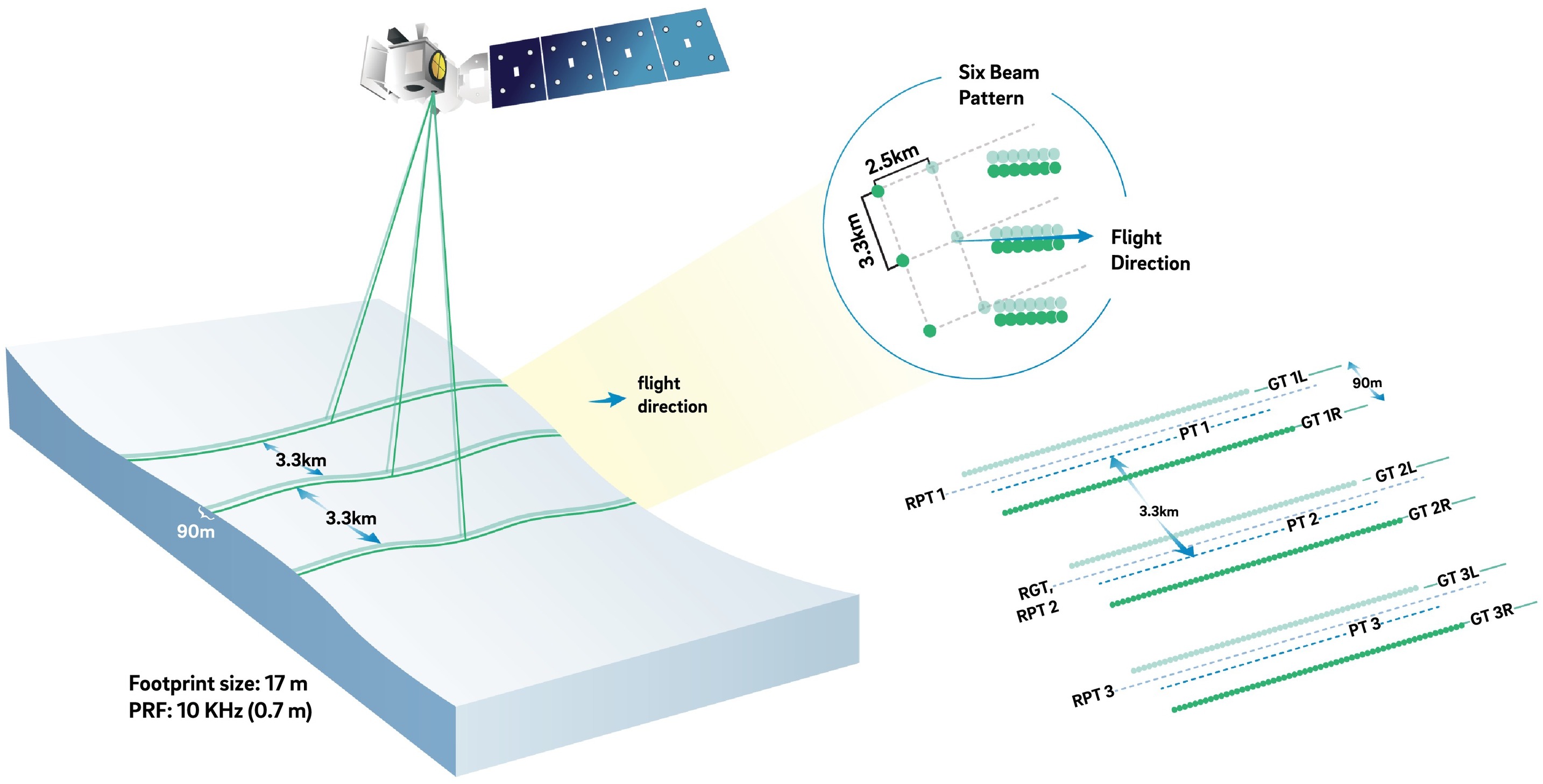
ICESat-2 laser configuration (from Smith and others, 2019)
We will use what we learned in the SWOT HR data access tutorial to compare the SWOT
ATL06:
- 17 m footprint
- Segments aggregate across 40 m of photos
- Segments acquired every 20 m along transect
To demonstrate a different earthaccess search method, we use a specific granule of SWOT data found using Earthdata search based on it being a high quality pass when we plot it. For comparison, we search for all ICESAT-2 tracks in a specified bounding box and manually pick one ICESat-2 track that intersects the SWOT swath at our region of interest.
Imports, paths, and constants to get started¶
# --- Pip Install a few packages that are not in the default CryoCloud environment: ---
%pip install -q pyTMD # For downloading and running the Circum-Antarctic Tidal Simulation (CATS) for tide corrections
%pip install -q icesat2_toolkit # For converting ICESat-2 time to GMT
%pip install -q cmap # For perceptually uniform colormaps
%pip install -q PyAstronomy # For date conversions
%pip install -q earthaccess==0.14Note: you may need to restart the kernel to use updated packages.
Note: you may need to restart the kernel to use updated packages.
Note: you may need to restart the kernel to use updated packages.
Note: you may need to restart the kernel to use updated packages.
Note: you may need to restart the kernel to use updated packages.
Best practice: %pip install library within notebook
%pip install library within notebookEnsures replicability for next person and that the library gets installed in the correct location
# --- Imports ---
%matplotlib widget
from pathlib import Path
from itertools import chain
from concurrent.futures import ProcessPoolExecutor
import datetime as dt
import os, logging, warnings, tempfile, gzip, shutil
# Data and processing
import h5py
import s3fs
import fiona
import h5coro
import rasterio
import requests
import earthaccess
import numpy as np
import pandas as pd
import xarray as xr
import icesat2_toolkit
from io import BytesIO
import geopandas as gpd
import cartopy.crs as ccrs
from PyAstronomy import pyasl
from scipy.interpolate import RegularGridInterpolator
import pyTMD
import timescale
# Plotting
import shapely
import shapefile
from shapely.plotting import plot_line
from shapely.geometry import LineString
from cmap import Colormap
import cartopy.crs as ccrs
from pyproj import CRS, Transformer
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib.ticker as ticker
from matplotlib.lines import Line2D
from matplotlib.patches import Patch
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
from mpl_toolkits.axes_grid1.anchored_artists import AnchoredSizeBar
# Quiet noisy logs
warnings.filterwarnings("ignore", message=".*ERROR parsing kml Style: No id.*", category=RuntimeWarning)
logging.getLogger("urllib3.connectionpool").setLevel(logging.ERROR)# --- Study area ---
DATADIR = Path("/home/jovyan/shared-public/SWOT/data") # Shared data files that cannot be streamed
GL_SHP = DATADIR / "Antarctica_masks/scripps_antarctica_polygons_v1.shp" # Grounding line shapefile
BBOX = [-1_970_000, 490_000, -1_820_000, 670_000] # PS71 [EPSG:3031] bounding box for Bach Ice Shelf
RIFT_BBOX = [-1_885_000, 570_000, -1_855_000, 590_000] # PS71 [EPSG:3031] bounding box for rift of interest
PS71 = ccrs.Stereographic(central_latitude=-90, central_longitude=0, true_scale_latitude=-71)
tick_label_size = 16
legend_label_size = 18# --- Earthdata login (interactive once; persists to ~/.netrc) ---
import earthaccess
auth = earthaccess.login(strategy="interactive", persist=True)
assert auth.authenticated, "Earthdata Login failed."# Coordinate transforms
def xy2ll(x, y):
"""Convert PS71 (EPSG:3031) coordinates to WGS84 (EPSG:4326)"""
crs_ll = CRS("EPSG:4326")
crs_xy = CRS("EPSG:3031")
xy_to_ll = Transformer.from_crs(crs_xy, crs_ll, always_xy=True)
lon, lat = xy_to_ll.transform(x, y)
return lon, lat
def ll2xy(lon, lat):
"""Convert WGS84 (EPSG:4326) coordinates to PS71 (EPSG:3031)"""
crs_ll = CRS("EPSG:4326")
crs_xy = CRS("EPSG:3031")
ll_to_xy = Transformer.from_crs(crs_ll, crs_xy, always_xy=True)
x, y = ll_to_xy.transform(lon, lat)
return x, y
def ps712utm(x, y, crs):
"""Convert UTM zone {crs} (SWOT native) to PS71 (EPSG:3031) coordinates"""
crs_utm = CRS(f"EPSG:{crs}")
crs_xy = CRS("EPSG:3031")
ll_to_xy = Transformer.from_crs(crs_xy, crs_utm, always_xy=True)
utmx, utmy = ll_to_xy.transform(x, y)
return utmx, utmy
# Colorbar Logic
def add_inset_colorbar(fig, ax, mappable, label,
anchor=(0.999, 0.90), # (x, y) in axes fraction
width=3.0, height=0.9, # inches
orientation="horizontal", extend=None, fontsize=tick_label_size):
# outer box (absolute size in inches, 2-tuple anchor is OK)
cbbox = inset_axes(ax, width=width, height=height,
bbox_to_anchor=anchor, bbox_transform=ax.transAxes, loc="upper right")
for sp in cbbox.spines.values():
sp.set_visible(False)
cbbox.set_facecolor([0, 0, 0, 0.7])
cbbox.set_xticks([]); cbbox.set_yticks([])
# actual bar inside
inner = inset_axes(cbbox, "92%", "20%", loc="center")
cbar = fig.colorbar(mappable, cax=inner, orientation=orientation, extend=extend)
cbar.outline.set_edgecolor("white"); cbar.outline.set_linewidth(1)
cbar.ax.tick_params(labelsize=fontsize, color="white", labelcolor="white")
cbar.set_label(label, fontsize=fontsize+2, color="white")
if orientation == "horizontal":
cbar.ax.xaxis.set_label_position("top")
cbar.ax.minorticks_on(); cbar.ax.tick_params(which="minor", length=4, color="white")
return cbarStreaming: No download needed!
Because these are in the right formats (.tif) and stored using the best data practices (no authentication, CAPTCHA, or other barriers), we can use python to quickly read this data directly into memory without the need to download.
# --- MOA 2009 grayscale basemap (gzipped GeoTIFF) ---
# URL of the protected gzip file
moa_url = "https://daacdata.apps.nsidc.org/pub/DATASETS/nsidc0593_moa2009_v02/geotiff/moa750_2009_hp1_v02.0.tif.gz"
# Use Earthdata login stored in ~/.netrc (standard for requests)
session = requests.Session()
session.auth = requests.utils.get_netrc_auth(moa_url)
# Add user-agent to mimic a real browser
headers = {"User-Agent": "NASA Earthdata Python client"}
# Download and check for success
response = session.get(moa_url, headers=headers)
response.raise_for_status()
# Confirm content-type is not HTML (debug print)
if b"<html" in response.content[:100].lower():
raise RuntimeError("Download returned HTML (login failed or wrong credentials?)")
# Decompress the GZip in memory
with gzip.GzipFile(fileobj=BytesIO(response.content)) as gz:
tif_bytes = BytesIO(gz.read())
# Read with Rasterio from memory
with rasterio.MemoryFile(tif_bytes) as memfile:
with memfile.open() as moa:
bounds = moa.bounds
left, bottom, right, top = bounds.left, bounds.bottom, bounds.right, bounds.top
moa_dat = moa.read(1)
ext = (left, right, bottom, top)WARNING:rasterio._env:CPLE_AppDefined in The definition of geographic CRS EPSG:4326 got from GeoTIFF keys is not the same as the one from the EPSG registry, which may cause issues during reprojection operations. Set GTIFF_SRS_SOURCE configuration option to EPSG to use official parameters (overriding the ones from GeoTIFF keys), or to GEOKEYS to use custom values from GeoTIFF keys and drop the EPSG code.
# --- SWOT HR granule (search via granule name) ---
search_days = 9
swot_results = earthaccess.search_data(
short_name="SWOT_L2_HR_Raster_D",
temporal=("2025-05-23", "2025-05-23"),
granule_name="SWOT_L2_HR_Raster_100m_UTM18D_N_x_x_x_033_143_012F_20250523T232533_20250523T232551_PID0_01.nc",
)
swot_file = earthaccess.open(swot_results)[0]
ds_swot = xr.open_dataset(swot_file, engine="h5netcdf")
# Build LAT/LON polygon for ATL06 search
poly_xy = np.array([[RIFT_BBOX[0], RIFT_BBOX[1]],[RIFT_BBOX[2], RIFT_BBOX[1]],
[RIFT_BBOX[2], RIFT_BBOX[3]],[RIFT_BBOX[0], RIFT_BBOX[3]],[RIFT_BBOX[0], RIFT_BBOX[1]]])
poly_lon, poly_lat = xy2ll(poly_xy[:,0], poly_xy[:,1])
polygon_ll = np.c_[poly_lon, poly_lat].tolist()
# Time window: SWOT start/end ± 9 days
t0 = dt.datetime.strptime(ds_swot.time_granule_start, "%Y-%m-%dT%H:%M:%S.%fZ")
t1 = dt.datetime.strptime(ds_swot.time_granule_end, "%Y-%m-%dT%H:%M:%S.%fZ")
atl06_window = (t0 - dt.timedelta(days=search_days), t1 + dt.timedelta(days=search_days))
atl06_results = earthaccess.search_data(short_name="ATL06",
temporal=atl06_window,
polygon=polygon_ll)
print(f'{len(atl06_results)} files found in time window in RIFT_BBOX')3 files found in time window in RIFT_BBOX
# --- Read a subset of ATL06 beams and variables - 30 s ---
beam_roots = ["gt1l","gt1r","gt2l","gt2r","gt3l","gt3r"]
s3_creds = auth.get_s3_credentials(daac="NSIDC")
fs_nsidc = s3fs.S3FileSystem(
key=s3_creds["accessKeyId"],
secret=s3_creds["secretAccessKey"],
token=s3_creds["sessionToken"],
)
def read_atl06_points(result):
"""Return a list of dicts (one per beam) with coords, heights, corrections."""
out = []
s3_url = result.data_links(access="direct")[0].replace("s3://","")
for beam in beam_roots:
try:
with fs_nsidc.open(s3_url, mode="rb", cache_type="blockcache", block_size=4*1024*1024) as s3obj:
ds_main = xr.open_dataset(s3obj, engine="h5netcdf",
group=f"{beam}/land_ice_segments",
driver_kwds={"rdcc_nbytes": 1024*1024},
decode_times=False, phony_dims="sort")
ds_dem = xr.open_dataset(s3obj, engine="h5netcdf",
group=f"{beam}/land_ice_segments/dem",
driver_kwds={"rdcc_nbytes": 1024*1024},
decode_times=False, phony_dims="sort")
ds_geo = xr.open_dataset(s3obj, engine="h5netcdf",
group=f"{beam}/land_ice_segments/geophysical",
driver_kwds={"rdcc_nbytes": 1024*1024},
decode_times=False, phony_dims="sort")
data = {
"lon": ds_main["longitude"].values,
"lat": ds_main["latitude"].values,
"h_li": ds_main["h_li"].values.astype("float32"),
"t_dt": ds_main["delta_time"].values,
"q": ds_main["atl06_quality_summary"].values,
"geoid_h": ds_dem["geoid_h"].values.astype("float32"),
"geoid_free2mean": ds_dem["geoid_free2mean"].values.astype("float32"),
"tide_ocean": ds_geo["tide_ocean"].values.astype("float32"),
"dac": ds_geo["dac"].values.astype("float32"),
"tide_earth": ds_geo["tide_earth"].values.astype("float32"),
"tide_earth_free2mean": ds_geo["tide_earth_free2mean"].values.astype("float32"),
"tide_pole": ds_geo["tide_pole"].values.astype("float32"),
"tide_load": ds_geo["tide_load"].values.astype("float32"),
"beam": beam, "name": s3_url,
}
# Mask poor quality data
bad = (data["q"] == 1) | (data["h_li"] > 3.0e38)
data["h_li"][bad] = np.nan
# PS71 coords
data["x"], data["y"] = ll2xy(data["lon"], data["lat"])
out.append(data)
except Exception as exc:
# Skip missing beams
continue
return out
atl06 = []
for r in atl06_results:
atl06.extend(read_atl06_points(r))
# Convert delta_time -> decimal years using icesat2_toolkit
for d in atl06:
dec = icesat2_toolkit.convert_delta_time(d["t_dt"])
d["time"] = dec["decimal"]
print(f'{len(atl06)} beams loaded')18 beams loaded
Preview and choose ATL06 tracks to compare with SWOT¶
We’ll plot a slice of the atl06 candidates on top of the SWOT swath and the MOA context map.
Use the is2_start/is2_end indices to step through candidates until you find a track that best crosses your area of interest (e.g., a crevasse field). The cell prints the granule filename and beam for each plotted candidate so you can note the one you want to use in the next step.
# --- Choose a slice of ATL06 candidates to preview (adjust as needed) ---
is2_start, is2_end = 0, 18 # tweak these indices
subset = atl06[is2_start:is2_end]
print(f"Previewing ATL06 candidates [{is2_start}:{is2_end}] out of total {len(atl06)}")
for d in subset:
print(f"{d['beam']:>4} {d['name'].split('/')[-1]}")
# --- Figure: MOA (gray) + SWOT swath (viridis) + ATL06 points ---
fig = plt.figure(figsize=(8, 8))
ax = plt.axes(projection=PS71)
# MOA basemap
ax.imshow(moa_dat, extent=ext, cmap="gray", vmin=15000, vmax=17000)
# SWOT swath (water surface elevation)
wse = ds_swot["wse"]
utm = ccrs.UTM(zone=ds_swot.utm_zone_num, southern_hemisphere=True)
X, Y = np.meshgrid(wse["x"].values, wse["y"].values)
mesh = ax.pcolormesh(
X, Y, wse.values,
transform=utm, cmap="viridis", shading="auto",
vmin=0, vmax=100, zorder=2
)
# ATL06 candidate tracks (colored by index within the slice)
cmap = plt.get_cmap("plasma")
n = max(1, len(subset))
for i, d in enumerate(subset):
color = cmap(i / (n - 1) if n > 1 else 0.5)
ax.scatter(d["x"], d["y"], s=2, color=color, transform=PS71, zorder=3)
# Convert cmap into ScalarMappable for colorbar
norm = plt.Normalize(vmin=0, vmax=n - 1)
sm = plt.cm.ScalarMappable(cmap=cmap, norm=norm)
sm.set_array([])
bach_rect = plt.Rectangle((RIFT_BBOX[0], RIFT_BBOX[1]), RIFT_BBOX[2] - RIFT_BBOX[0], RIFT_BBOX[3] - RIFT_BBOX[1],
zorder=5, linewidth=2, edgecolor="white", facecolor="none")
ax.add_patch(bach_rect)
# View & labels
ax.set_xlim(BBOX[0], BBOX[2])
ax.set_ylim(BBOX[1], BBOX[3])
ax.set_title("ATL06 candidate tracks over SWOT swath (Bach Ice Shelf)", fontsize=14)
# Colorbar for SWOT Swath
add_inset_colorbar(fig, ax, mesh, "SWOT WSE [m]", anchor=(0.999, 0.87), extend="max")
# ICESat-2 tracks colorbar
add_inset_colorbar(fig, ax, sm, "ICESat-2 Index", anchor=(0.999, 0.999))
plt.tight_layout()
plt.show()Previewing ATL06 candidates [0:18] out of total 18
gt1l ATL06_20250515130244_09102710_007_01.h5
gt1r ATL06_20250515130244_09102710_007_01.h5
gt2l ATL06_20250515130244_09102710_007_01.h5
gt2r ATL06_20250515130244_09102710_007_01.h5
gt3l ATL06_20250515130244_09102710_007_01.h5
gt3r ATL06_20250515130244_09102710_007_01.h5
gt1l ATL06_20250520014201_09792712_007_01.h5
gt1r ATL06_20250520014201_09792712_007_01.h5
gt2l ATL06_20250520014201_09792712_007_01.h5
gt2r ATL06_20250520014201_09792712_007_01.h5
gt3l ATL06_20250520014201_09792712_007_01.h5
gt3r ATL06_20250520014201_09792712_007_01.h5
gt1l ATL06_20250524013336_10402712_007_01.h5
gt1r ATL06_20250524013336_10402712_007_01.h5
gt2l ATL06_20250524013336_10402712_007_01.h5
gt2r ATL06_20250524013336_10402712_007_01.h5
gt3l ATL06_20250524013336_10402712_007_01.h5
gt3r ATL06_20250524013336_10402712_007_01.h5
# --- Find granule matching ATL06_20241213201922_13522510_006_01.h5 ---
gran = 'ATL06_20250515130244_09102710_007_01.h5'
beam = 'gt2r'
is2_tracks_to_use = []
for i, data in enumerate(atl06):
if gran in data['name'] and beam in data['beam']:
is2_tracks_to_use.append(atl06[i])
assert is2_tracks_to_use, 'No tracks found' # Check at least one track chosen# --- Stream MeASUREs ice velocity data via earthaccess (NSIDC-0484), 450 m grid ---
v_results = earthaccess.search_data(short_name="NSIDC-0484",
cloud_hosted=True,
temporal=("1996-01-01","1996-01-02"),
count=1)
v_file = earthaccess.open(v_results)[0]
vel = xr.open_dataset(v_file)
# Crop a margin around our box; compute magnitude for plotting
off = 10_000
v_crop = vel.sel(x=slice(BBOX[0]-off, BBOX[2]+off),
y=slice(BBOX[3]+off, BBOX[1]-off))
vel_mag = np.hypot(v_crop.VX, v_crop.VY) # m/yr
oslo = Colormap("crameri:oslo").to_matplotlib() # Velocity colormap# --- Advect ICESat-2 to location at time of SWOT data collection ---
# Create regular grid interpolators to speed up running the advection code (~10x speed up over using native grid)
def prep_velocity_interpolators(vel):
vx_interp = RegularGridInterpolator((vel.y.values[::-1], vel.x.values),
vel["VX"].values[::-1, :],bounds_error=False,fill_value=np.nan)
vy_interp = RegularGridInterpolator((vel.y.values[::-1], vel.x.values),
vel["VY"].values[::-1, :],bounds_error=False,fill_value=np.nan)
return vx_interp, vy_interp
for track in is2_tracks_to_use:
# Copy arrays so we don't mutate the originals
start_x = track["x"].copy()
start_y = track["y"].copy()
start_time = track["time"].copy() # decimal years (one per point)
# Spatial mask to the study bounding box (BBOX: [xmin, ymin, xmax, ymax])
mask = (
(start_x > BBOX[0]) &
(start_x < BBOX[2]) &
(start_y > BBOX[1]) &
(start_y < BBOX[3])
)
# Keep full array length; mark points outside the box as NaN in X
# (Y is left unchanged to preserve indexing; NaNs in X will propagate during updates)
start_x[~mask] = np.nan
# Convert decimal year → Python datetime for each sample
start_time = [pyasl.decimalYearGregorianDate(t) for t in start_time]
# Integration setup
target_time = t0 - dt.timedelta(days=0) # SWOT target time (t0); minus 0 keeps semantics
step_size = 1 / 365 # years per step (1 day)
# For each point, compute time difference to target, then number of steps
steps = [(target_time - t) for t in start_time] # list of timedeltas (can be ±)
steps = [int( td.total_seconds() / (365 * 24 * 60 * 60) / step_size ) for td in steps]
# Use the sign of the FIRST point's step count to set direction (forward or backward in time)
step_size = np.sign(steps[0]) * step_size
# Integrate a UNIFORM number of steps for all points (assumes similar times)
steps = np.abs(steps)
# Velocity interpolators on the cropped grid
# v_crop has coordinates (y, x). Arrays are stored row-major with y decreasing,
# so the helper flips y when building RegularGridInterpolator.
vx_interp, vy_interp = prep_velocity_interpolators(v_crop)
# Euler integration (meters per year * years per step = meters per step)
x = start_x.copy()
y = start_y.copy()
for _ in range(steps[0]): # same number of steps for every point
# Sample velocity at current positions; inputs are (y, x)
vx = vx_interp((y, x)) # meters / year
vy = vy_interp((y, x))
# Update positions; NaNs remain NaN and will propagate
x = x + vx * step_size
y = y + vy * step_size
# Store advected coordinates back on the track
track["x_advected"] = x
track["y_advected"] = yApply comparable geophysics & interpolate SWOT along-track¶
We bring heights to a consistent reference, then interpolate the SWOT wse at the advected ICESat-2 points (in the granule’s UTM zone).
# --- Ensure CATS2008-v2023 is available locally for PyTMD ---
fs = s3fs.S3FileSystem(anon=True)
pyTMD_path = pyTMD.utilities.get_data_path("data")
model = pyTMD.io.model(pyTMD_path, verify=False).elevation("CATS2008-v2023")
model.grid_file.parent.mkdir(parents=True, exist_ok=True)
if not model.grid_file.exists():
with fs.open("pytmd/CATS2008_v2023/CATS2008_v2023.nc", "rb") as fin, \
open(model.grid_file, "wb") as fout:
shutil.copyfileobj(fin, fout)
model.grid_file.exists()True# Tides
def compute_tides(model, lons, lats, datetimes):
"""
Computes the tides at given locations and times using PyTMD.
Parameters
model: PyTMD.model
Tide model
lons: list[float]
Geodetic longitude in EPSG:4326
lats: list[float]
Geodetic latitude in EPSG:4326
datetimes: list[datetime.datetime]
Datetime to calculate tides
Returns
tides - list[float]
Tide elevations in cm at locations and times sepcified
"""
years = np.array([d.year for d in datetimes])
months = np.array([d.month for d in datetimes])
days = np.array([d.day for d in datetimes])
hours = np.array([d.hour for d in datetimes])
minutes= np.array([d.minute for d in datetimes])
tide_time = timescale.time.convert_calendar_dates(years, months, days, hours, minutes)
# Setup model
constituents = pyTMD.io.OTIS.read_constants(model.grid_file, model.model_file,
model.projection, type=model.type,
grid=model.format, apply_flexure=False)
c = constituents.fields
amp, ph, _ = pyTMD.io.OTIS.interpolate_constants(np.atleast_1d(lons),
np.atleast_1d(lats), constituents,
type=model.type, method="spline",
extrapolate=True)
cph = -1j * ph * np.pi / 180.0
hc = amp * np.exp(cph)
# Calculate tides and infer minor constitutents at each location and time
tides = []
for i in range(len(datetimes)):
TIDE = pyTMD.predict.map(tide_time[i], hc, c, deltat=0, corrections=model.format)
MINOR = pyTMD.predict.infer_minor(tide_time[i], hc, c, deltat=0, corrections=model.format)
t_cm = (TIDE.data + MINOR.data) * 100.0
tides.append(t_cm.astype("float32"))
return np.array(tides) # --- Find tide corrections using CATS at times of interest - 1m 29s ---
# Swot and ICESat-2 times
tide_times = [
t0,
pyasl.decimalYearGregorianDate(is2_tracks_to_use[0]["time"][0]),
]
sats = ["SWOT", "IS2"]
data = is2_tracks_to_use[0]
data["lon_advected"], data["lat_advected"] = xy2ll(data["x_advected"], data["y_advected"])
for tide_time, satellite in zip(tide_times, sats):
tide_results = np.array(
compute_tides(model, data["lon_advected"], data["lat_advected"], [tide_time])).T
data[f"tide_{satellite}"] = tide_results
data["tide_IS2"] = data["tide_IS2"].astype("float32").squeeze()
data["tide_SWOT"] = data["tide_SWOT"].astype("float32").squeeze()# --- Apply geophysical corrections to ICESat-2 heights for fair comparison to SWOT ---
data["h_li_corrected"] = (
data["h_li"]
- data["geoid_h"]
+ data["geoid_free2mean"]
- data["tide_earth"]
- data["tide_pole"]
- data["tide_load"]
)# --- Interpolate SWOT swath along icesat-2 track ---
# Create arrays to hold interpolated values
overall_wse = np.zeros_like(data["x_advected"])
overall_wse[:] = np.nan
overall_wse_sigma = np.zeros_like(data["x_advected"])
overall_wse_sigma[:] = np.nan
wse = ds_swot["wse"]
wse_sigma = ds_swot["wse_uncert"]
# Convert icesat2 to utm, and interpolate SWOT advected data
x_utm = wse["x"].values
y_utm = wse["y"].values
advected_x_utm, advected_y_utm = ps712utm(data["x_advected"], data["y_advected"], utm.to_epsg())
advected_x_utm = xr.DataArray(advected_x_utm, dims=["points"])
advected_y_utm = xr.DataArray(advected_y_utm, dims=["points"])
x_utm = wse["x"].values
y_utm = wse["y"].values
interpolated_wse = wse.interp(
x=advected_x_utm,
y=advected_y_utm,
method="linear",
kwargs={"fill_value": np.nan},
).compute()
interpolated_wse_sigma = wse_sigma.interp(
x=advected_x_utm,
y=advected_y_utm,
method="linear",
kwargs={"fill_value": np.nan},
).compute()
#Put all non-nan values into overall_wse
# If there are no SWOT data values where we have IS2 data, add a nan
mask = np.isnan(overall_wse) & ~np.isnan(interpolated_wse)
# Only update at those places
overall_wse[mask] = interpolated_wse[mask]
overall_wse_sigma[mask] = interpolated_wse_sigma[mask]
# Compute +- 2 std
lower = overall_wse - 2 * overall_wse_sigma
upper = overall_wse + 2 * overall_wse_sigma
# Cropping data to the rift plot area
min_mask = -1872000
max_mask = -1865000
x_mask = (data["x_advected"] > min_mask) & (
data["x_advected"] < max_mask
)# --- Apply tide corrections to both datasets and uncertainty bounds ---
data["is2_tidecorrected"] = data["h_li_corrected"] - data["tide_IS2"] / 100
data["swot_tidecorrected"] = overall_wse - data["tide_SWOT"] / 100
data["swot_lower"] = lower - data["tide_SWOT"] / 100
data["swot_upper"] = upper - data["tide_SWOT"] / 100# Plot comparison
label = "Elevation [m]"
vmin = 0
vmax = 50
fig = plt.figure(figsize=(8, 12))
utm = ccrs.UTM(zone=ds_swot.utm_zone_num, southern_hemisphere=True)
ps71_projection = ccrs.Stereographic(central_latitude=-90, central_longitude=0, true_scale_latitude=-71)
gs = gridspec.GridSpec(2, 1, height_ratios=[4, 1])
ax = fig.add_subplot(gs[0], projection=ps71_projection)
# Map View
ax.imshow(moa_dat, extent=ext, cmap="gray", vmin=15000, vmax=17000)
cb = ax.imshow(vel_mag, cmap=oslo,
extent=[vel_mag["x"].min(), vel_mag["x"].max(), vel_mag["y"].min(), vel_mag["y"].max()],
origin="upper", alpha=0.6, vmin=0, vmax=550)
grounded_3031.plot(ax=ax, transform=ps71, color="none", edgecolor="white", linewidth=1, zorder=4)
wse = ds_swot['wse']
x = wse["x"].values
y = wse["y"].values
X, Y = np.meshgrid(x, y)
mesh = ax.pcolormesh(X, Y, wse.values, transform=utm, cmap="viridis", shading="auto", vmin=vmin, vmax=vmax)
ax.scatter(data["x_advected"], data["y_advected"], color="white", s=5, transform=ps71_projection)
x_plotting = data["x_advected"][x_mask]
y_plotting = data["y_advected"][x_mask]
ax.scatter(x_plotting, y_plotting, color="orangered", s=5, transform=ps71_projection)
# Cross section
ax2 = fig.add_subplot(gs[1])
ax2.plot(data["x_advected"], data["is2_tidecorrected"],
color="orangered", linewidth=2, label="ICESat-2")
ax2.plot(data["x_advected"], data["swot_tidecorrected"],
color="navy", linewidth=2, label="SWOT")
ax2.fill_between(data["x_advected"], data["swot_lower"],
data["swot_upper"], color="navy", alpha=0.3)
# SWOT Swath colorbar
add_inset_colorbar(fig, ax, mesh, label="SWOT WSE [m]", anchor=(0.999, 0.87), extend="max")
# Ice Velocity colorbar
add_inset_colorbar(fig, ax, cb, label="Ice Velocity [m/a]", anchor=(0.999, 0.999))
# Prettify panel 1
ax.set_xlim(BBOX[0] + 1000, BBOX[2] - 1000)
ax.set_ylim(BBOX[1] + 1000, BBOX[3] - 1000)
# Scalebar
scalebar = AnchoredSizeBar(ax.transData, 20000, "20 km", "lower right", pad=0.1,
sep=1, color="white", frameon=False, size_vertical=2000,
fontproperties=mpl.font_manager.FontProperties(size=20, weight="bold"), label_top=False)
# Antarctica Inset
inset = fig.add_axes([0.15, 0.73, 0.3, 0.3], projection=ps71_projection) # [left, bottom, width, height]
inset.patch.set_facecolor("none")
for spine in inset.spines.values():
spine.set_visible(False)
inset.set_xticks([])
inset.set_yticks([])
grounded_3031.plot(ax=inset, transform=ps71, color="lightgray", edgecolor="lightgray", linewidth=0.3, zorder=1)
shelves_3031.plot(ax=inset, transform=ps71, color="white", edgecolor="white", linewidth=0.3, zorder=2)
rect = plt.Rectangle((BBOX[0], BBOX[1]), BBOX[2] - BBOX[0], BBOX[3] - BBOX[1],
zorder=3, linewidth=2, edgecolor="red", facecolor="none")
inset.add_patch(rect)
ax.add_artist(scalebar)
scalebar.set_bbox_to_anchor((0.99, 0.67), transform=ax.transAxes)
# Time Labels
ax.text(0.01, 0.01, "SWOT: 2025-05-23\nICESat-2: 2025-05-15", transform=ax.transAxes, fontsize=legend_label_size, color="white")
# Polish panel 2
ax2.set_xlabel("PS71 X [km]", fontsize=legend_label_size)
ax2.set_ylabel("Elevation [m]", fontsize=legend_label_size)
KM_SCALE = 1e3
ticks_x = ticker.FuncFormatter(lambda x, pos: "{0:g}".format(x / KM_SCALE))
ax2.xaxis.set_major_formatter(ticks_x)
ax2.tick_params(axis="both", which="major", labelsize=tick_label_size, size=10)
ax2.spines["top"].set_visible(False)
ax2.spines["right"].set_visible(False)
ax2.set_xlim(np.nanmin(data["x_advected"]), np.nanmax(data["x_advected"]))
ax2.set_xlim(min_mask, max_mask)
ax2.set_ylim(-2, 32)
# Legend
handles = [
Line2D([0], [0], color="orangered", linewidth=2, label="ICESat-2"),
Line2D([0], [0], color="navy", linewidth=2, label="SWOT"),
Patch(facecolor="navy", alpha=0.3, label="SWOT Uncertainty"),
]
ax2.legend(fontsize=legend_label_size, loc="lower left", handles=handles[:1] + [handles[1]])
leg = ax2.get_legend()
for line in leg.get_lines():
line.set_linewidth(3)
leg.get_frame().set_edgecolor("white")
leg.get_frame().set_linewidth(1)
fig.tight_layout()- Depoorter, M. A., Bamber, J. L., Griggs, J., Lenaerts, J. T. M., Ligtenberg, S. R. M., van den Broeke, M. R., & Moholdt, G. (2013). Antarctic masks (ice-shelves, ice-sheet, and islands), link to shape file. PANGAEA. 10.1594/PANGAEA.819147
- Smith, B., Fricker, H. A., Holschuh, N., Gardner, A. S., Adusumilli, S., Brunt, K. M., Csatho, B., Harbeck, K., Huth, A., Neumann, T., Nilsson, J., & Siegfried, M. R. (2019). Land ice height-retrieval algorithm for NASA’s ICESat-2 photon-counting laser altimeter. Remote Sensing of Environment, 233, 111352. 10.1016/j.rse.2019.111352